PrimerStation is a web service to calculate optimal primer sets that are guaranteed high specificity against whole human genome.
To achieve high accuracy, we used hybridization ratio of primers in the liquid solution. Calculating the status of the sequence hybridization in terms of stringent hybridization ratio is computational costly, there was no web service that checks whole human genome and returns highly specific primer set which calculated by the precise physicochemical model.
To shorten the response time, we precomputed the candidates of the specific primers by using a massively parallel computer with 100 CPUs (SunFire 15K) for about 2 months in advance. This enables PrimerStation to search and output the qualified primers interactively.
PrimerStation has a capability of selecting highly specific primers suitable for multiplex PCR by seeking optimal temperature range that minimizes the possibility of cross-reaction. It also allows users to add some heuristic rules in the primer design, e.g., exclusion of SNPs in primers and avoidance of poly-A and CA-repeats in PCR products.
We performed substantial tests that verified the PCR amplification of randomly selected primers for ChrX, and we confirmed that the primers perfectly magnify specific PCR products. The details of the method will be reported elsewhere. We believe PrimerStation must be beneficial to a wide range of biologists and medical scientists.
To design genomic PCR primers, you need to specify target region by using the form of genomic region or RefSeq gene ID. When you design multiplex genomic PCR primers, you are required to input every genomic region that you are going to amplify all together. You can input the regions delimited by commas, spaces, or new lines. The number of multiple inputs is limited up to 40 genomic regions.
| Chromosomal range | chrchromosome:start-end (e.g., chr20:180000-190000) |
|---|---|
| The region of the target genomic sequence can be specified by the above syntax. You can use 'chr1'-'chr22','chrX' or 'chrY' as chromosome. The genomic region starting from start to end is used as the amplification target. | |
| RefSeq gene identifiers | accession[@exon number] (e.g., NM_003380@2) |
| RefSeq gene accession number accession possibly with an exon number. | |
|
NM_003380 chr20:180000-190000 chr12:2600000-2700000 |
The input example to design three multiplex genomic PCR primers on three genomic regions; the region specified by the RefSeq identifier NM_003380, the chromosome 20 range from 180000 to 190000, and the chromosome 12 range from 2600000 to 2700000.
|
NM_003380 NM_003380 NM_003380 chr20:180000-190000 chr20:180000-190000 |
The input example to design five multiplex genomic PCR primers for three different products on the identical region of mRNA with NM_003380 and two distinct amplicons on the same chromosomal region on chromosome 20 range from 180000 to 190000.
| NM_003380@1, NM_003380@3, NM_003380@8, |
The input example to design three primer pairs on the first, second, and eighth exons of the same region with NM_003380. You can specify the three regions by using comma delimited expression.
In PrimerStation, you can design primers with various parameters including the range of product size, the minimum product size difference for multiplex primers, cation concentration, primer concentration, whether you need to avoid designing primers with known SNPs, undesirable secondary structures, PCR product with (A)n or (CA)n repeats.
When you design genomic PCR primers, the selection of the product size is crucial. To detect all of the primers with electrophoresis at once, we used the region of the product size from 60bp to 600bp. These parameters are set as default parameters. If you are unable to design the specified region with the parameters, you may find more candidates by expanding the size of region.
This parameter is used for designing multiplex genomic primers to avoid designing primers which product size are similar. If the product sizes of designed primers are similar, it is difficult to distinguish which region is amplified by the primers. For the sake of simplicity, we can set the minimum product size difference. PrimerStation avoid designing primers that have similar product size within the specified threshold. In other words, you can set the minimum PCR product size differences between any PCR products.
If you design primers on SNPs, the primers may not amplify the target sequence perfectly. PrimerStation can avoid the potential failure of the hybridization exerting the best efforts. By checking the check box, PrimerStation avoid designing primers with known SNPs. The SNP information is retrieved from dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/).
The secondary structure on the primer sequence is one of the reasons of unsuccessful primer design. PrimerStation avoid designing primers with undesirable secondary structures. The most stable secondary structures and its free energy changes are calculated by Mfold. If the secondary structure is stable, the free energy change becomes small. We can specify the threshold of the minimum free energy changes by selecting the number in the select box. This step requires certain amount of calculation time.
PolyA ,polyT, (CA)n or (GT)n repeats in the PCR products trigger slippage of amplification which is the cause of unintended size of products. To avoid such slippage, you need to avoid designing primers which amplify products with polyA, polyT, (CA)n, (GT)n repeats by checking checkbox of each corresponding option.
The result of primer design is shown as following figure.
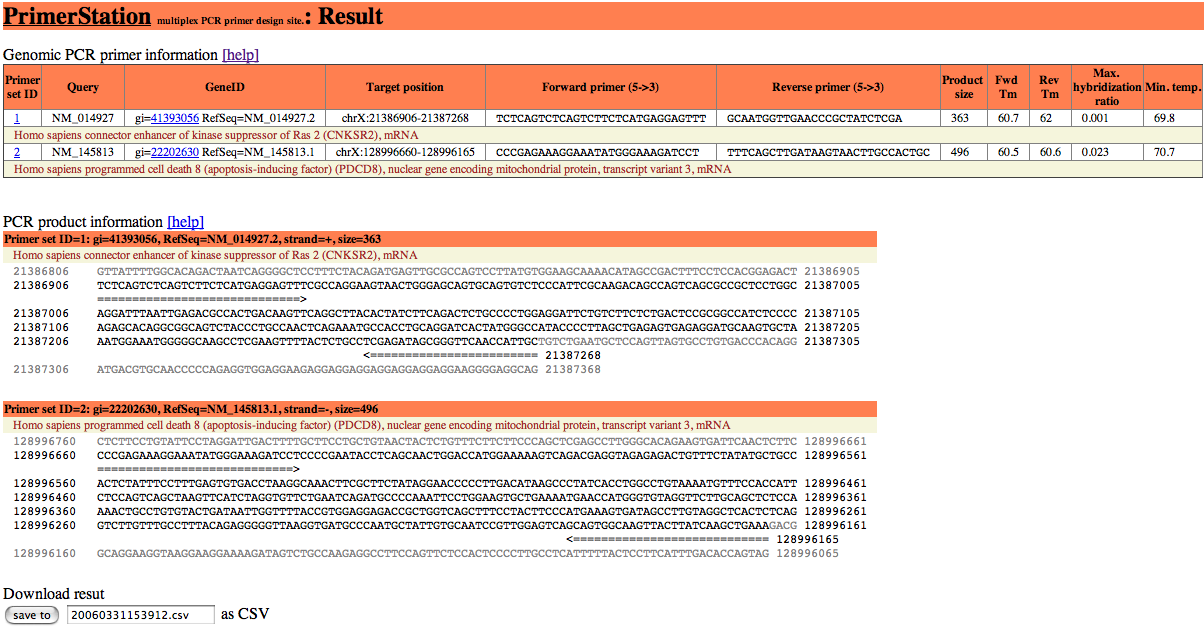
The result is shown in two separated sections. The first section indicates the primer information and the second section is the product information. The results can be downloaded as in CSV file format
This section consists of following data; primer set ID, user's query, gene ID, target position, forward primer sequence, reverse primer sequence, product size, Tm for forward primer, Tm for reverse primer, maximum value for the second maximum hybridization ratio, minimum executable temperature. If you selected the option to calculate secondary structure, PrimerStation returns the result of secondary structure calculation.
The detail description for the primer information is as follows:
| Primer set ID | Serial number for target sequences. |
|---|---|
| Query | Query sequence which was submitted by user. |
| GeneID | GenBank ID (e.g., gi=41393056) and RefSeq ID (e.g., RefSeq=NM=014927.2) for specified gene.GenBank ID (e.g., gi=41393056) and RefSeq ID (e.g., RefSeq=NM=014927.2) for specified gene. |
| Target position | Target position of the designed primer set. |
| Forward primer | Sequence of the forward primer. |
| Reverse primer | Sequence of the reverse primer. |
| Product size | Product size for designed primer set. |
| Fwd Tm | Tm of forward primer calculated based on nearest-neighbor model. |
| Rev Tm | Tm of reverse primer calculated based on nearest-neighbor model. |
| Max. hybridization ratio | Maximum hybridization ratio between the primer and its possible off-targets. |
| Min. temp. | Minimum annealing temperature at which the primers have sufficient specificity. |
This section provides information of product sequences that are wedged between primers and external sides of the product. The product sequence is shown as black characters and the external sides of the product are displayed as grey characters. The primer set is depicted by the arrowed line.
You can download the result for your design in csv (comma sepalated values) format. The csv file can be read by Microsoft Excel.
All accesses to PrimerStation are saved and processed to take out statistics of accesses, query patterns, and heavy users. Information provided by users would be used to enhance the function of PrimerStation. Frequent accesses from unresolved IP addreses or proxy server can be set as the object of refusal.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas (C) and the Leading Project for Biosimulation from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a joint research project with SONY Corporation. Computational time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science and the University of Tokyo.